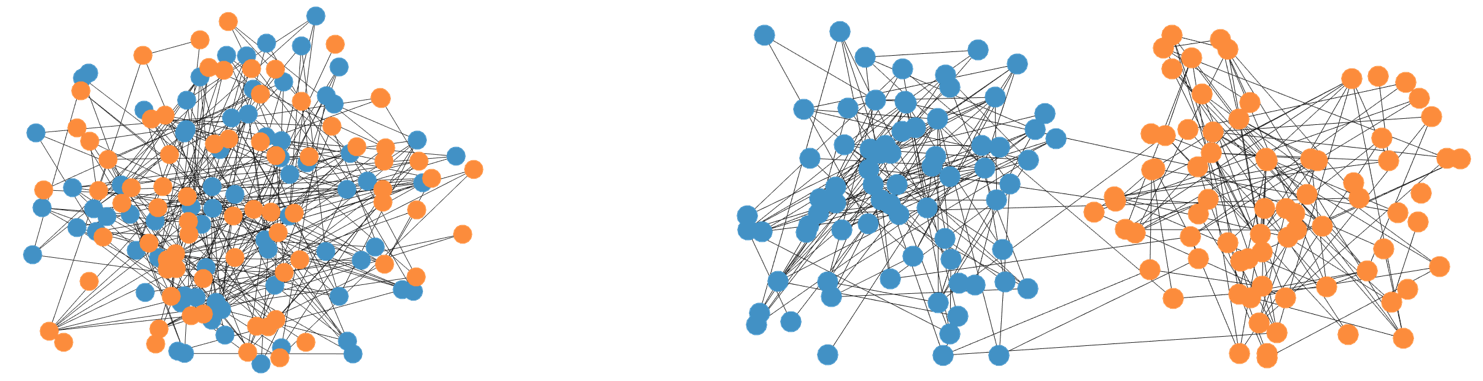
Spectral Stitching
A community-recovery approach for haplotype phasing
Haplotype Phasing
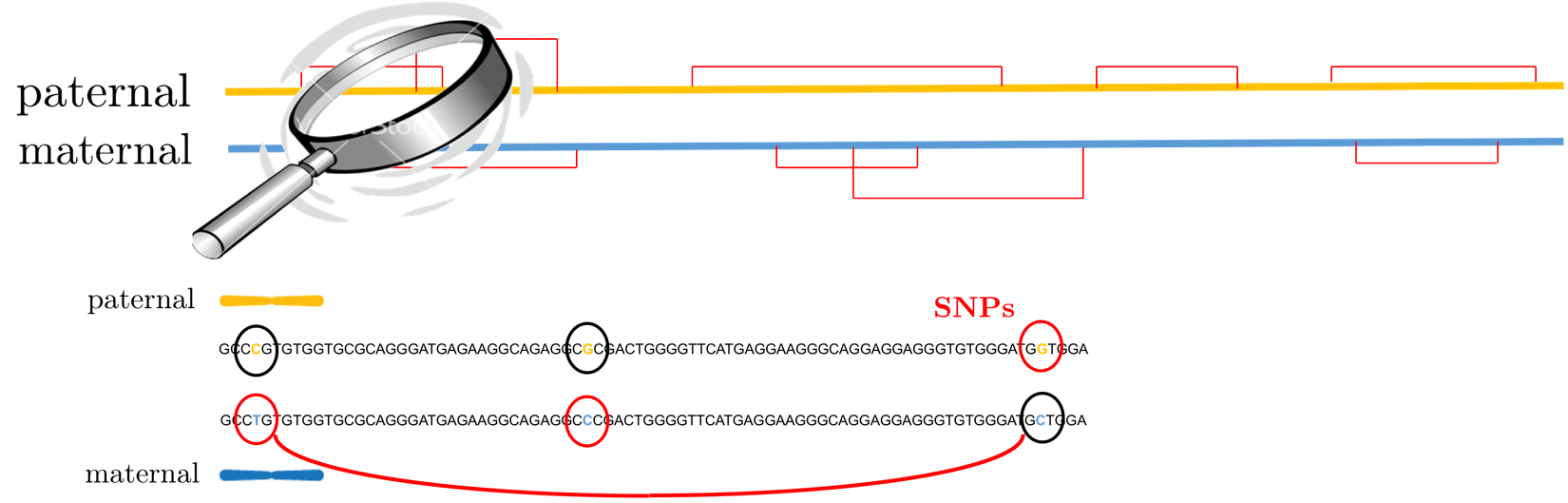
Humans have 23 pairs of homologous chromosomes, one maternal and one paternal. Each pair are identical sequences of nucleotides A,G,C,T’s except on certain documented positions called single nucleotide polymorphisms (SNPs), or genetic variants. At each of these positions, one of the chromosomes takes on one of A,G,C or T which is the same as the majority of the population (called the major allele), while the other chromosome takes on a variant (also called minor allele). A haplotype of an individual is the pair of sequences of SNPs on the two homologous chromosomes. Knowing the haplotypes of individuals can lead to a better understanding of the interplay of genetic variation and disease as well as better inference of human demographic history. The haplotype phasing problem aims at inferring haplotypes of individuals from high-throughput sequencing data.
Connection to Community Detection
The advent of next generation sequencing technologies allows haplotype phasing by providing linking reads between multiple SNP locations. The linking reads give noisy measurements of whether two (or more) SNPs have the variant on the same chromosome or different chromosomes. One can formulate the problem of haplotype phasing as recovery of two communities of SNP locations — those with the variant on the maternal chromosome and those with the variant on the paternal chromosome — from a collection of linking reads. This is very similar to the community recovery problem in statistics and computer science. The challenge comes from a sample locality issue: the linking reads come mainly from nearby SNPs rather than uniformly sampled between all node pairs, as in most existing theory.